2. Joe Model Batch Run
2026-02-03
Source:vignettes/a02-joe-model-batch-run.Rmd
a02-joe-model-batch-run.RmdOverview
This tutorial demonstrates how to run the Joe Model in batch mode to evaluate multiple management or climate scenarios simultaneously. Building on Tutorial 1: Joe Model Overview, you will learn how to: - Create and structure a scenarios file - Run the Joe Model across multiple scenarios in a single function call - Visualize and compare results across scenarios and locations
Prerequisites
Before starting this tutorial, you should: - Complete Tutorial 1: Joe Model Overview -
Understand how stressor-response and stressor-magnitude workbooks are
structured - Have basic familiarity with ggplot2 for
visualization
Why Use Batch Runs?
In real-world applications, you often need to compare multiple scenarios to inform management decisions. Common use cases include:
| Use Case | Example |
|---|---|
| Climate projections | Compare current conditions vs. 2050 vs. 2080 climate scenarios |
| Recovery actions | Evaluate the benefits of different restoration strategies |
| Sensitivity analysis | Test how changes in specific stressors affect outcomes |
| Risk assessment | Compare best-case, expected, and worst-case scenarios |
Rather than running the Joe Model separately for each scenario and manually combining results, the batch function automates this process and returns a consolidated dataset ready for analysis.
How Batch Runs Work
The batch run function works by modifying stressor magnitudes according to multipliers you specify in a scenarios file:
- Base condition: The original stressor-magnitude values serve as your baseline
- Scenario multipliers: Each scenario specifies which stressors to modify and by how much
- Automatic iteration: The function loops through all scenarios, adjusting stressor values and running the Joe Model
- Consolidated output: Results from all scenarios are combined into a single data frame
For example, a multiplier of 1.2 increases a stressor by
20%, while 0.8 decreases it by 20%.
Understanding the Scenarios File
The scenarios file is a simple Excel workbook (or data frame) that defines what changes to apply in each scenario. Here’s an example structure:

Scenarios File Structure
The scenarios file has a specific format:
| Column | Description |
|---|---|
| Scenario | Unique name for the scenario (e.g., “climate_2050”, “restore_riparian”) |
| Stressor columns | One column per stressor you want to modify, containing the multiplier values |
Key points: - Each row represents one scenario -
Stressor column names must exactly match the names in
your stressor-magnitude workbook - Values are
multipliers: 1.0 = no change,
1.2 = +20%, 0.5 = -50% - Leave cells blank or
use 1.0 for stressors that shouldn’t change in that
scenario - You only need to include columns for stressors you want to
modify
Example Scenarios
Here are some example scenario configurations:
| Scenario | Temperature | Sediment | Flow_alteration | Interpretation |
|---|---|---|---|---|
| baseline | 1.0 | 1.0 | 1.0 | Current conditions (no changes) |
| climate_moderate | 1.1 | 1.0 | 0.9 | 10% warmer, 10% less flow |
| climate_severe | 1.25 | 1.2 | 0.7 | 25% warmer, 20% more sediment, 30% less flow |
| restoration | 0.9 | 0.6 | 1.0 | Riparian restoration reduces temp and sediment |
Import Data
The batch run requires three input files:
- Stressor-magnitude workbook: Baseline stressor values at each location
- Stressor-response workbook: Dose-response relationships for each stressor
- Scenarios file: Multipliers defining each scenario (see above)
# ---------------------------------------------------------
# 1. Import stressor-magnitude workbook
# This contains the baseline stressor values at each location
# ---------------------------------------------------------
filename_sm <- system.file("extdata", "stressor_magnitude_unc_ARTR.xlsx", package = "CEMPRA")
stressor_magnitude <- StressorMagnitudeWorkbook(
filename = filename_sm,
scenario_worksheet = "natural_unc"
)
# ---------------------------------------------------------
# 2. Import stressor-response workbook
# This defines how each stressor affects system capacity
# ---------------------------------------------------------
filename_sr <- system.file("extdata", "stressor_response_fixed_ARTR.xlsx", package = "CEMPRA")
stressor_response <- StressorResponseWorkbook(filename = filename_sr)
# ---------------------------------------------------------
# 3. Import the scenarios file
# This defines the multipliers for each scenario
# ---------------------------------------------------------
filename_sc <- system.file("extdata", "scenario_batch_joe.xlsx", package = "CEMPRA")
scenarios <- readxl::read_excel(filename_sc)Let’s examine the scenarios file to understand what scenarios we’re comparing:
# View the scenarios configuration
print(scenarios)
#> # A tibble: 8 × 4
#> Scenario Stressor Metric Multiplier
#> <chr> <chr> <chr> <dbl>
#> 1 scenario_1 Temperature_adult Mean 1.2
#> 2 scenario_1 Temperature_adult SD 1.1
#> 3 scenario_2 Aug_flow Mean 0.7
#> 4 scenario_2 Aug_flow SD 1.1
#> 5 scenario_3 Nat_lim_other Mean 0.5
#> 6 scenario_4 Temperature_adult Mean 0.8
#> 7 scenario_4 Nat_lim_other Mean 0.8
#> 8 scenario_4 Aug_flow Mean 0.8Run the Joe Model in Batch Mode
Now we run the Joe Model across all scenarios. The
JoeModel_Run_Batch() function:
- Reads the baseline stressor magnitudes
- For each scenario, applies the specified multipliers to adjust stressor values
- Runs the Joe Model with Monte Carlo simulation
- Combines all results into a single data frame
# Run the Joe Model in batch mode across all scenarios
# Note: MC_sims is set low (3) for this tutorial - use higher values (100+)
# for real applications to get stable uncertainty estimates
exp_dat <- JoeModel_Run_Batch(
scenarios = scenarios, # Scenario definitions with multipliers
dose = stressor_magnitude, # Baseline stressor magnitudes
sr_wb_dat = stressor_response, # Stressor-response relationships
MC_sims = 3 # Number of Monte Carlo simulations per scenario
)Understanding the Output
The output exp_dat is a data frame with one row per
location × scenario × Monte Carlo replicate:
# View the first few rows of results
head(exp_dat, 6)
#> # A tibble: 6 × 4
#> # Groups: HUC, simulation [6]
#> HUC simulation CE Scenario
#> <int> <int> <dbl> <chr>
#> 1 1701010201 1 0 scenario_1
#> 2 1701010201 2 0 scenario_1
#> 3 1701010201 3 0 scenario_1
#> 4 1701010202 1 0.582 scenario_1
#> 5 1701010202 2 0.584 scenario_1
#> 6 1701010202 3 0.436 scenario_1
# Check the dimensions
cat("\nTotal rows:", nrow(exp_dat), "\n")
#>
#> Total rows: 1236
cat("Unique scenarios:", length(unique(exp_dat$Scenario)), "\n")
#> Unique scenarios: 4
cat("Unique locations (HUCs):", length(unique(exp_dat$HUC)), "\n")
#> Unique locations (HUCs): 103| Column | Description |
|---|---|
| HUC | Location identifier (Hydrologic Unit Code or custom ID) |
| Scenario | Which scenario this result belongs to |
| simulation | Monte Carlo replicate number |
| CE | Cumulative Effects score (System Capacity, 0-1 scale) |
Visualizing Results
With batch results, there are two natural ways to visualize the data:
- By location: Compare all scenarios for a single watershed
- By scenario: Compare all watersheds within a single scenario
Interpreting System Capacity Scores
Before plotting, it’s helpful to understand how to interpret system capacity values:
| Score Range | Interpretation | Color Code |
|---|---|---|
| 0.75 - 1.00 | Good: High habitat capacity, minimal cumulative stress | Green |
| 0.50 - 0.75 | Moderate: Some degradation, recovery actions may help | Yellow |
| 0.20 - 0.50 | Poor: Significant stress, likely population impacts | Orange |
| 0.00 - 0.20 | Critical: Severe degradation, urgent action needed | Pink/Red |
These thresholds are general guidelines; appropriate thresholds may vary by species and system.
Plot 1: Compare Scenarios for a Single Location
This visualization answers: “How do different scenarios affect system capacity at a specific watershed?”
This is useful for: - Presenting results to local stakeholders - Evaluating which recovery actions would benefit a priority watershed - Comparing climate impact projections for a specific location
# ---------------------------------------------------------
# Select a single watershed to visualize
# Replace this HUC with one relevant to your study area
# ---------------------------------------------------------
target_huc <- 1701010204
huc_subset <- exp_dat[exp_dat$HUC == target_huc, ]
# ---------------------------------------------------------
# Calculate summary statistics across Monte Carlo replicates
# Mean gives the central estimate; SD shows uncertainty
# ---------------------------------------------------------
huc_summary <- huc_subset %>%
group_by(Scenario) %>%
summarise(
mean = mean(CE, na.rm = TRUE),
sd = sd(CE, na.rm = TRUE),
.groups = "drop"
) %>%
mutate(
lower = mean - sd,
upper = mean + sd
)
# ---------------------------------------------------------
# Create the plot with color-coded capacity zones
# ---------------------------------------------------------
ggplot(huc_summary, aes(x = mean, y = Scenario, xmin = lower, xmax = upper)) +
# Add background zones for capacity interpretation
geom_rect(aes(xmin = 0, xmax = 0.2, ymin = -Inf, ymax = Inf),
fill = "pink", alpha = 0.02) +
geom_rect(aes(xmin = 0.2, xmax = 0.5, ymin = -Inf, ymax = Inf),
fill = "orange", alpha = 0.02) +
geom_rect(aes(xmin = 0.5, xmax = 0.75, ymin = -Inf, ymax = Inf),
fill = "yellow", alpha = 0.02) +
geom_rect(aes(xmin = 0.75, xmax = 1, ymin = -Inf, ymax = Inf),
fill = "green", alpha = 0.02) +
# Add points and error bars
geom_point(size = 3) +
geom_errorbarh(height = 0.2) +
# Labels and styling
ggtitle(paste("Watershed:", target_huc)) +
xlab("System Capacity (0 - 1)") +
ylab("Scenario") +
scale_x_continuous(limits = c(0, 1)) +
theme_bw() +
theme(
plot.title = element_text(hjust = 0.5),
panel.grid.minor = element_blank()
)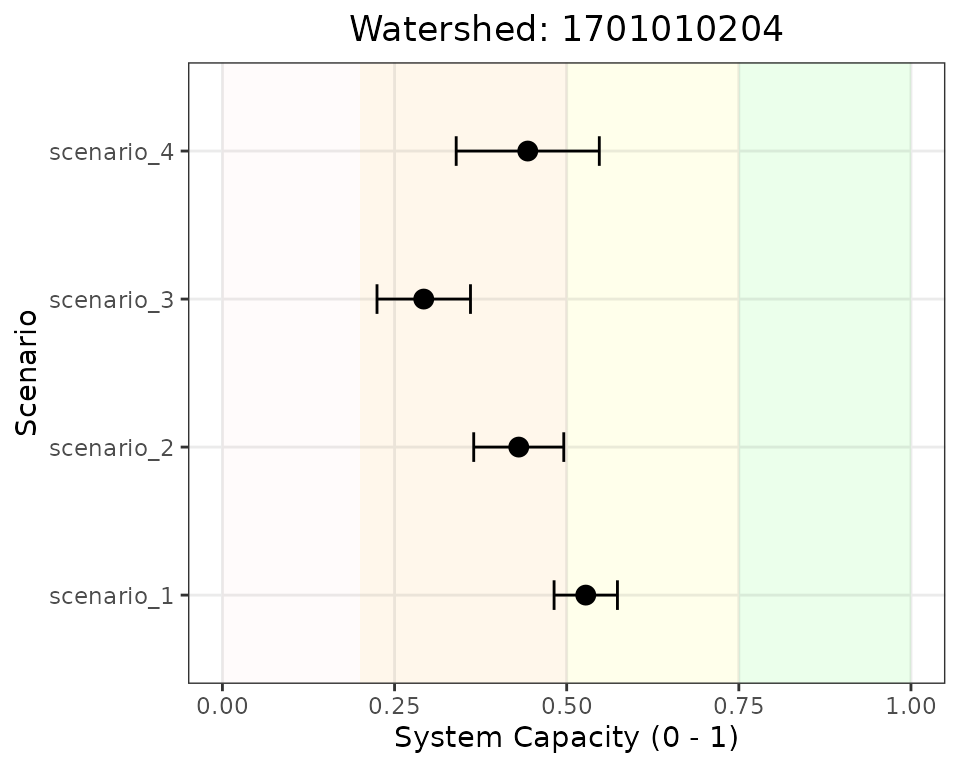
System capacity across scenarios for a single watershed. Error bars show standard deviation from Monte Carlo simulations.
Reading this plot: - Each point shows the mean system capacity for that scenario - Error bars indicate uncertainty (±1 standard deviation from Monte Carlo simulation) - Background colors indicate capacity zones (green = good, pink = critical) - Scenarios can be compared by their horizontal position and overlap
Plot 2: Compare Locations Within a Single Scenario
This visualization answers: “Which watersheds are most/least affected under a given scenario?”
This is useful for: - Prioritizing locations for management action - Identifying spatial patterns of vulnerability - Reporting landscape-wide results for a specific scenario
# ---------------------------------------------------------
# Select a single scenario to visualize
# ---------------------------------------------------------
target_scenario <- "scenario_1"
scenario_subset <- exp_dat[exp_dat$Scenario == target_scenario, ]
# ---------------------------------------------------------
# Calculate summary statistics for each watershed
# ---------------------------------------------------------
scenario_summary <- scenario_subset %>%
group_by(HUC) %>%
summarise(
mean = mean(CE, na.rm = TRUE),
sd = sd(CE, na.rm = TRUE),
.groups = "drop"
) %>%
mutate(
lower = mean - sd,
upper = mean + sd,
HUC = as.character(HUC) # Convert to character for proper y-axis display
) %>%
# Sort by mean capacity for easier interpretation
arrange(desc(mean))
# Reorder factor levels for sorted display
scenario_summary$HUC <- factor(scenario_summary$HUC, levels = rev(scenario_summary$HUC))
# ---------------------------------------------------------
# Create the plot
# ---------------------------------------------------------
ggplot(scenario_summary, aes(x = mean, y = HUC, xmin = lower, xmax = upper)) +
# Add background zones
geom_rect(aes(xmin = 0, xmax = 0.2, ymin = -Inf, ymax = Inf),
fill = "pink", alpha = 0.02) +
geom_rect(aes(xmin = 0.2, xmax = 0.5, ymin = -Inf, ymax = Inf),
fill = "orange", alpha = 0.02) +
geom_rect(aes(xmin = 0.5, xmax = 0.75, ymin = -Inf, ymax = Inf),
fill = "yellow", alpha = 0.02) +
geom_rect(aes(xmin = 0.75, xmax = 1, ymin = -Inf, ymax = Inf),
fill = "green", alpha = 0.02) +
# Add points and error bars
geom_point(size = 2) +
geom_errorbarh(height = 0.3) +
# Labels and styling
ggtitle(paste("Scenario:", target_scenario)) +
xlab("System Capacity (0 - 1)") +
ylab("Watershed (HUC)") +
scale_x_continuous(limits = c(0, 1)) +
theme_bw() +
theme(
plot.title = element_text(hjust = 0.5),
panel.grid.minor = element_blank(),
axis.text.y = element_text(size = 7) # Smaller text for many watersheds
)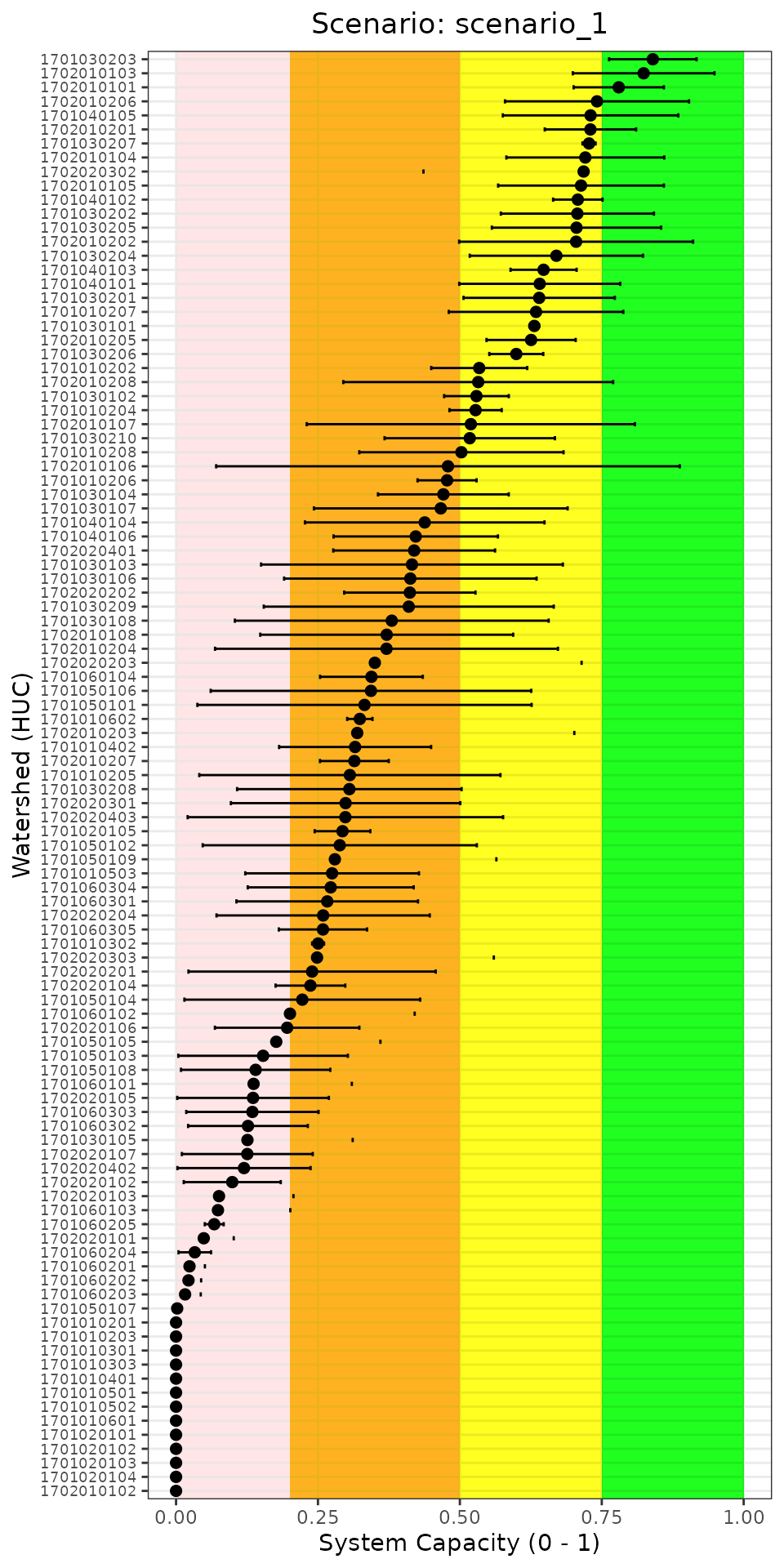
System capacity across all watersheds for a single scenario. This view helps identify which locations are most vulnerable.
Reading this plot: - Watersheds are sorted from highest to lowest capacity (top to bottom) - Watersheds in the pink/orange zones may be priorities for restoration - Wide error bars indicate high uncertainty, possibly due to variable stressor data
Advanced: Comparing Scenario Differences
Beyond visualizing raw scores, you may want to quantify how much each scenario changes capacity relative to baseline. This helps identify which scenarios have the largest positive or negative impact.
# ---------------------------------------------------------
# Calculate mean capacity per scenario (across all locations)
# ---------------------------------------------------------
scenario_means <- exp_dat %>%
group_by(Scenario) %>%
summarise(
mean_capacity = mean(CE, na.rm = TRUE),
sd_capacity = sd(CE, na.rm = TRUE),
min_capacity = min(CE, na.rm = TRUE),
max_capacity = max(CE, na.rm = TRUE),
.groups = "drop"
) %>%
arrange(desc(mean_capacity))
print(scenario_means)
#> # A tibble: 4 × 5
#> Scenario mean_capacity sd_capacity min_capacity max_capacity
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 scenario_1 0.335 0.281 0 0.916
#> 2 scenario_2 0.327 0.214 0 0.753
#> 3 scenario_4 0.299 0.223 0 0.811
#> 4 scenario_3 0.204 0.170 0 0.739This summary table shows: - mean_capacity: Average system capacity across all locations and Monte Carlo runs - sd_capacity: Variability in capacity scores - min/max_capacity: The range of outcomes observed
Saving Results for Further Analysis
You can export the batch results for use in other analyses or reporting:
# ---------------------------------------------------------
# Export full results to CSV
# ---------------------------------------------------------
write.csv(exp_dat, "joe_model_batch_results.csv", row.names = FALSE)
# ---------------------------------------------------------
# Export scenario summary statistics
# ---------------------------------------------------------
write.csv(scenario_means, "scenario_summary.csv", row.names = FALSE)Tips for Your Application
Choosing Multiplier Values
When creating your scenarios file, consider:
- Use empirical data when available: Climate projections, monitoring data, or modeled outputs provide defensible multipliers
- Document your assumptions: Record the rationale for each multiplier value
- Test sensitivity: Try a range of multipliers to understand how sensitive results are to your assumptions
- Consider interactions: A scenario with multiple stressor changes may have non-linear cumulative effects
Increasing Monte Carlo Simulations
For publication or decision-making, increase MC_sims to
get stable estimates:
# Recommended: 100+ simulations for stable uncertainty estimates
exp_dat <- JoeModel_Run_Batch(
scenarios = scenarios,
dose = stressor_magnitude,
sr_wb_dat = stressor_response,
MC_sims = 100 # Increase for real applications
)Next Steps
Now that you can run batch scenarios with the Joe Model, consider:
| Tutorial | Description |
|---|---|
| 3. Population Model Overview | Link system capacity to population dynamics |
| 4. Population Model Batch Run | Run population projections across scenarios |
| 5. BC CEF Data Import | Import data from BC Cumulative Effects Framework |
| 6. Population Model Evaluation | Sensitivity analysis and model evaluation |
For additional guidance, see the CEMPRA Documentation.